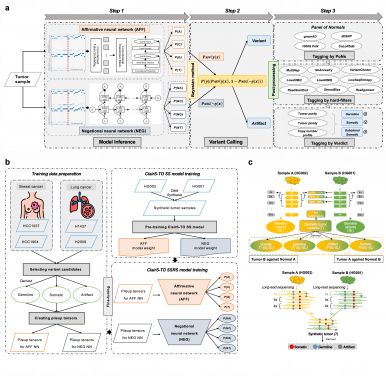
Researchers at The University of Hong Kong have introduced two advanced deep-learning algorithms, ClairS-TO and Clair3-RNA, enhancing genetic mutation detection crucial for cancer diagnostics and RNA studies. Led by Professor Ruibang Luo, the innovative tools utilize long-read sequencing technologies to boost mutation detection accuracy, paving the way for better precision medicine and genomic research. Published in Nature Communications, ClairS-TO eliminates the need for matched healthy tissue samples, facilitating reliable tumor analysis even with scarce materials. Clair3-RNA, the first deep-learning variant caller for long-read RNA sequencing, effectively distinguishes real mutations from noise, allowing comprehensive analysis of gene expression and mutations. Part of the renowned Clair series, these open-source algorithms have over 400,000 downloads globally, solidifying their place in computational biology. This research heralds a new era in accessible and accurate genetic analysis, aiming to improve cancer diagnoses and enhance personalized medicine, benefiting patients and researchers alike.
Read more about the studies: ClairS-TO | Clair3-RNA.